VERB(DATA_TO_TRANSFORM, STUFF_IT_DOES)Amounts and proportions
Lesson for Monday, June 10, 2024–Friday, June 14, 2024
When you visualize proportions with ggplot, you’ll typically go through a two-step process:
- Summarize the data with {dplyr} (typically with a combination of
group_by()andsummarize()) - Plot the summarized data
To help with the intuition behind the dplyr verbs mutate(), group_by(), and summarize(), go look at these animations, and come back to them throughout the class.
Manipulating data with {dplyr}
You had some experience with {dplyr} functions in the RStudio primers, but we’ll briefly review them here.
There are 6 important verbs that you’ll typically use when working with data:
- Extract rows/cases with
filter() - Extract columns/variables with
select() - Arrange/sort rows with
arrange() - Make new columns/variables with
mutate() - Make group summaries with
group_by |> summarize()
Every {dplyr} verb follows the same pattern. The first argument is always a data frame, and the function always returns a data frame:
Filtering with filter()
The filter() function takes two arguments: a data frame to transform, and a set of tests. It will return each row for which the test is TRUE.
This code, for instance, will look at the gapminder dataset and return all rows where country is equal to “Denmark”:
filter(gapminder, country == "Denmark")
## # A tibble: 12 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Denmark Europe 1952 70.8 4334000 9692.
## 2 Denmark Europe 1957 71.8 4487831 11100.
## 3 Denmark Europe 1962 72.4 4646899 13583.
## 4 Denmark Europe 1967 73.0 4838800 15937.
## 5 Denmark Europe 1972 73.5 4991596 18866.
## 6 Denmark Europe 1977 74.7 5088419 20423.
## 7 Denmark Europe 1982 74.6 5117810 21688.
## 8 Denmark Europe 1987 74.8 5127024 25116.
## 9 Denmark Europe 1992 75.3 5171393 26407.
## 10 Denmark Europe 1997 76.1 5283663 29804.
## 11 Denmark Europe 2002 77.2 5374693 32167.
## 12 Denmark Europe 2007 78.3 5468120 35278.Notice that there are two equal signs (==). This is because it’s a logical test, similar to greater than (>) or less than (<). When you use a single equal sign, you set an argument (like data = gapminder); when you use two, you are doing a test. There are lots of different ways to do logical tests:
| Test | Meaning |
|---|---|
x < y |
Less than |
x > y |
Greater than |
x == y |
Equal to |
x <= y |
Less than or equal to |
x >= y |
Greater than or equal to |
x != y |
Not equal to |
x %in% y |
In (group membership) |
is.na(x) |
Is missing |
!is.na(x) |
Is not missing |
Use filter() and logical tests to show:
- The data for Canada
- All data for countries in Oceania
- Rows where life expectancy is greater than 82
# The data for Canada
filter(gapminder, country == "Canada")
# All data for countries in Oceania
filter(gapminder, continent == "Oceania")
# Rows where life expectancy is greater than 82
filter(gapminder, lifeExp > 82)You can also use multiple conditions, and these will extract rows that meet every test. By default, if you separate the tests with a comma, R will consider this an “and” test and find rows that are both Denmark and greater than 2000.
filter(gapminder, country == "Denmark", year > 2000)
## # A tibble: 2 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Denmark Europe 2002 77.2 5374693 32167.
## 2 Denmark Europe 2007 78.3 5468120 35278.You can also use “or” with “|” and “not” with “!”:
| Operator | Meaning |
|---|---|
a & b |
and |
a | b |
or |
!a |
not |
Use filter() and logical tests to show:
- Canada before 1970
- Countries where life expectancy in 2007 is below 50
- Countries where life expectancy in 2007 is below 50 and are not in Africa
# Canada before 1970
filter(gapminder, country == "Canada", year < 1970)
# Countries where life expectancy in 2007 is below 50
filter(gapminder, year == 2007, lifeExp < 50)
# Countries where life expectancy in 2007 is below 50 and are not in Africa
filter(gapminder, year == 2007, lifeExp < 50, continent != "Africa")Beware of some common mistakes! You can’t collapse multiple tests into one. Instead, use two separate tests:
# This won't work!
filter(gapminder, 1960 < year < 1980)
# This will work
filter(gapminder, 1960 < year, year < 1980)Also, you can avoid stringing together lots of tests by using the %in% operator, which checks to see if a value is in a list of values.
# This works, but is tedious
filter(gapminder,
country == "Mexico" | country == "Canada" | country == "United States")
# This is more concise and easier to add other countries later
filter(gapminder,
country %in% c("Mexico", "Canada", "United States"))Adding new columns with mutate()
You create new columns with the mutate() function. You can create a single column like this:
mutate(gapminder, gdp = gdpPercap * pop)
## # A tibble: 1,704 × 7
## country continent year lifeExp pop gdpPercap gdp
## <fct> <fct> <int> <dbl> <int> <dbl> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779. 6567086330.
## 2 Afghanistan Asia 1957 30.3 9240934 821. 7585448670.
## 3 Afghanistan Asia 1962 32.0 10267083 853. 8758855797.
## 4 Afghanistan Asia 1967 34.0 11537966 836. 9648014150.
## 5 Afghanistan Asia 1972 36.1 13079460 740. 9678553274.
## 6 Afghanistan Asia 1977 38.4 14880372 786. 11697659231.
## 7 Afghanistan Asia 1982 39.9 12881816 978. 12598563401.
## 8 Afghanistan Asia 1987 40.8 13867957 852. 11820990309.
## 9 Afghanistan Asia 1992 41.7 16317921 649. 10595901589.
## 10 Afghanistan Asia 1997 41.8 22227415 635. 14121995875.
## # ℹ 1,694 more rowsAnd you can create multiple columns by including a comma-separated list of new columns to create:
mutate(gapminder, gdp = gdpPercap * pop,
pop_mill = round(pop / 1000000))
## # A tibble: 1,704 × 8
## country continent year lifeExp pop gdpPercap gdp pop_mill
## <fct> <fct> <int> <dbl> <int> <dbl> <dbl> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779. 6567086330. 8
## 2 Afghanistan Asia 1957 30.3 9240934 821. 7585448670. 9
## 3 Afghanistan Asia 1962 32.0 10267083 853. 8758855797. 10
## 4 Afghanistan Asia 1967 34.0 11537966 836. 9648014150. 12
## 5 Afghanistan Asia 1972 36.1 13079460 740. 9678553274. 13
## 6 Afghanistan Asia 1977 38.4 14880372 786. 11697659231. 15
## 7 Afghanistan Asia 1982 39.9 12881816 978. 12598563401. 13
## 8 Afghanistan Asia 1987 40.8 13867957 852. 11820990309. 14
## 9 Afghanistan Asia 1992 41.7 16317921 649. 10595901589. 16
## 10 Afghanistan Asia 1997 41.8 22227415 635. 14121995875. 22
## # ℹ 1,694 more rowsYou can also do conditional tests within mutate() using the ifelse() function. This works like the =IFELSE function in Excel. Feed the function three arguments: (1) a test, (2) the value if the test is true, and (3) the value if the test is false:
ifelse(TEST, VALUE_IF_TRUE, VALUE_IF_FALSE)We can create a new column that is a binary indicator for whether the country’s row is after 1960:
mutate(gapminder, after_1960 = ifelse(year > 1960, TRUE, FALSE))
## # A tibble: 1,704 × 7
## country continent year lifeExp pop gdpPercap after_1960
## <fct> <fct> <int> <dbl> <int> <dbl> <lgl>
## 1 Afghanistan Asia 1952 28.8 8425333 779. FALSE
## 2 Afghanistan Asia 1957 30.3 9240934 821. FALSE
## 3 Afghanistan Asia 1962 32.0 10267083 853. TRUE
## 4 Afghanistan Asia 1967 34.0 11537966 836. TRUE
## 5 Afghanistan Asia 1972 36.1 13079460 740. TRUE
## 6 Afghanistan Asia 1977 38.4 14880372 786. TRUE
## 7 Afghanistan Asia 1982 39.9 12881816 978. TRUE
## 8 Afghanistan Asia 1987 40.8 13867957 852. TRUE
## 9 Afghanistan Asia 1992 41.7 16317921 649. TRUE
## 10 Afghanistan Asia 1997 41.8 22227415 635. TRUE
## # ℹ 1,694 more rowsWe can also use text labels instead of TRUE and FALSE:
mutate(gapminder,
after_1960 = ifelse(year > 1960, "After 1960", "Before 1960"))
## # A tibble: 1,704 × 7
## country continent year lifeExp pop gdpPercap after_1960
## <fct> <fct> <int> <dbl> <int> <dbl> <chr>
## 1 Afghanistan Asia 1952 28.8 8425333 779. Before 1960
## 2 Afghanistan Asia 1957 30.3 9240934 821. Before 1960
## 3 Afghanistan Asia 1962 32.0 10267083 853. After 1960
## 4 Afghanistan Asia 1967 34.0 11537966 836. After 1960
## 5 Afghanistan Asia 1972 36.1 13079460 740. After 1960
## 6 Afghanistan Asia 1977 38.4 14880372 786. After 1960
## 7 Afghanistan Asia 1982 39.9 12881816 978. After 1960
## 8 Afghanistan Asia 1987 40.8 13867957 852. After 1960
## 9 Afghanistan Asia 1992 41.7 16317921 649. After 1960
## 10 Afghanistan Asia 1997 41.8 22227415 635. After 1960
## # ℹ 1,694 more rowsUse mutate() to:
- Add an
africacolumn that is TRUE if the country is on the African continent - Add a column for logged GDP per capita
- Add an
africa_asiacolumn that says “Africa or Asia” if the country is in Africa or Asia, and “Not Africa or Asia” if it’s not
# Add an africa column that is TRUE if the country is on the African continent
mutate(gapminder, africa = continent == "Africa")
# Add a column for logged GDP per capita
mutate(gapminder, log_gdpPercap = log(gdpPercap))
# Add an africa_asia column that says “Africa or Asia” if the country is in
# Africa or Asia, and “Not Africa or Asia” if it’s not
mutate(
gapminder,
africa_asia =
ifelse(continent %in% c("Africa", "Asia"), "Africa or Asia", "Not Africa or Asia")
)Combining multiple verbs with pipes (|>)
What if you want to filter to include only rows from 2002 and make a new column with the logged GDP per capita? Doing this requires both filter() and mutate(), so we need to find a way to use both at once.
One solution is to use intermediate variables for each step:
gapminder_2002_filtered <- filter(gapminder, year == 2002)
gapminder_2002_logged <- mutate(gapminder_2002_filtered, log_gdpPercap = log(gdpPercap))That works fine, but your environment panel will start getting full of lots of intermediate data frames.
Another solution is to nest the functions inside each other. Remember that all {dplyr} functions return data frames, so you can feed the results of one into another:
filter(mutate(gapminder, log_gdpPercap = log(gdpPercap)),
year == 2002)That works too, but it gets really complicated once you have even more functions, and it’s hard to keep track of which function’s arguments go where. I’d avoid doing this entirely.
See this short “Pipes” guide to learn about R’s two different styles of pipe: |> and %>%
One really nice solution is to use a pipe, or |>. The pipe takes an object on the left and passes it as the first argument of the function on the right.
# gapminder will automatically get placed in the _____ spot
gapminder |> filter(_____, country == "Canada")These two lines of code do the same thing:
filter(gapminder, country == "Canada")
gapminder |> filter(country == "Canada")Using pipes, you can start with a data frame, pass it to one verb, then pass the output of that verb to the next verb, and so on. When reading any code with a |>, it’s easiest to read the |> as “and then”. This would read:
Take the
gapminderdataset and then filter it so that it only has rows from 2002 and then add a new column with the logged GDP per capita
gapminder |>
filter(year == 2002) |>
mutate(log_gdpPercap = log(gdpPercap))Here’s another way to think about pipes more conceptually. This isn’t valid R code, obviously, but imagine you’re going to take yourself, and then wake up, get out of bed, get dressed, and leave the house. Writing that whole process as nested functions would look like this:
leave_house(get_dressed(get_out_of_bed(wake_up(me, time = "8:00"), side = "correct"), pants = TRUE, shirt = TRUE), car = TRUE, bike = FALSE)Instead of nesting everything, we can use pipes to chain these together. This would read
Take myself, and then wake up at 8:00, and then get out of bed on the correct side, and then get dressed with pants and a shirt, and then leave the house in a car
me |>
wake_up(time = "8:00") |>
get_out_of_bed(side = "correct") |>
get_dressed(pants = TRUE, shirt = TRUE) |>
leave_house(car = TRUE, bike = FALSE)Summarizing data by groups with group_by() |> summarize()
The summarize() verb takes an entire frame and calculates summary information about it. For instance, this will find the average life expectancy for the whole gapminder data:
gapminder |> summarize(mean_life = mean(lifeExp))
## # A tibble: 1 × 1
## mean_life
## <dbl>
## 1 59.5You can also make multiple summary variables, just like mutate(), and it will return a column for each:
gapminder |> summarize(mean_life = mean(lifeExp),
min_life = min(lifeExp))
## # A tibble: 1 × 2
## mean_life min_life
## <dbl> <dbl>
## 1 59.5 23.6Use summarize() to calculate:
- The first (minimum) year in the
gapminderdataset - The last (maximum) year in the dataset
- The number of rows in the dataset (use the {dplyr} cheatsheet)
- The number of distinct countries in the dataset (use the {dplyr} cheatsheet)
gapminder |>
summarize(first = min(year),
last = max(year),
num_rows = n(),
num_unique = n_distinct(country))Use filter() and summarize() to calculate the median life expectancy on the African continent in 2007:
gapminder |>
filter(continent == "Africa", year == 2007) |>
summarize(n_countries = n_distinct(country),
med_le = median(lifeExp))Notice that summarize() on its own summarizes the whole dataset, so you only get a single row back. These values are the averages and minimums for the entire data frame. If you group your data into separate subgroups, you can use summarize() to calculate summary statistics for each group. Do this with group_by().
The group_by() function puts rows into groups based on values in a column. If you run this:
gapminder |> group_by(continent)
## # A tibble: 1,704 × 6
## # Groups: continent [5]
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779.
## 2 Afghanistan Asia 1957 30.3 9240934 821.
## 3 Afghanistan Asia 1962 32.0 10267083 853.
## 4 Afghanistan Asia 1967 34.0 11537966 836.
## 5 Afghanistan Asia 1972 36.1 13079460 740.
## 6 Afghanistan Asia 1977 38.4 14880372 786.
## 7 Afghanistan Asia 1982 39.9 12881816 978.
## 8 Afghanistan Asia 1987 40.8 13867957 852.
## 9 Afghanistan Asia 1992 41.7 16317921 649.
## 10 Afghanistan Asia 1997 41.8 22227415 635.
## # ℹ 1,694 more rows…you won’t see anything different! R has put the dataset into separate invisible groups behind the scenes, but you haven’t done anything with those groups, so nothing has really happened. If you do things with those groups with summarize(), though, group_by() becomes much more useful.
For instance, this will take the gapminder data frame, group it by continent, and then summarize it by calculating the number of distinct countries in each group. It will return one row for each group, so there should be a row for each continent:
gapminder |>
group_by(continent) |>
summarize(n_countries = n_distinct(country))
## # A tibble: 5 × 2
## continent n_countries
## <fct> <int>
## 1 Africa 52
## 2 Americas 25
## 3 Asia 33
## 4 Europe 30
## 5 Oceania 2You can calculate multiple summary statistics, as before:
gapminder |>
group_by(continent) |>
summarize(n_countries = n_distinct(country),
avg_life_exp = mean(lifeExp))
## # A tibble: 5 × 3
## continent n_countries avg_life_exp
## <fct> <int> <dbl>
## 1 Africa 52 48.9
## 2 Americas 25 64.7
## 3 Asia 33 60.1
## 4 Europe 30 71.9
## 5 Oceania 2 74.3Find the minimum, maximum, and median life expectancy for each continent:
gapminder |>
group_by(continent) |>
summarize(min_le = min(lifeExp),
max_le = max(lifeExp),
med_le = median(lifeExp))Find the minimum, maximum, and median life expectancy for each continent in 2007 only:
gapminder |>
filter(year == 2007) |>
group_by(continent) |>
summarize(min_le = min(lifeExp),
max_le = max(lifeExp),
med_le = median(lifeExp))Finally, you can group by multiple columns and R will create subgroups for every combination of the groups and return the number of rows of combinations. For instance, we can calculate the average life expectancy by both year and continent and we’ll get 60 rows, since there are 5 continents and 12 years (5 × 12 = 60):
gapminder |>
group_by(continent, year) |>
summarize(avg_life_exp = mean(lifeExp))
## # A tibble: 60 × 3
## # Groups: continent [5]
## continent year avg_life_exp
## <fct> <int> <dbl>
## 1 Africa 1952 39.1
## 2 Africa 1957 41.3
## 3 Africa 1962 43.3
## 4 Africa 1967 45.3
## 5 Africa 1972 47.5
## 6 Africa 1977 49.6
## 7 Africa 1982 51.6
## 8 Africa 1987 53.3
## 9 Africa 1992 53.6
## 10 Africa 1997 53.6
## # ℹ 50 more rowsSelecting with select()
The last two verbs are far simpler than filter(), mutate(), and group_by() |> summarize().
You can choose specific columns with the select() verb. This will only keep two columns: lifeExp and year:
gapminder |> select(lifeExp, year)
## # A tibble: 1,704 × 2
## lifeExp year
## <dbl> <int>
## 1 28.8 1952
## 2 30.3 1957
## 3 32.0 1962
## 4 34.0 1967
## 5 36.1 1972
## 6 38.4 1977
## 7 39.9 1982
## 8 40.8 1987
## 9 41.7 1992
## 10 41.8 1997
## # ℹ 1,694 more rowsYou can remove specific columns by prefacing the column names with -, like -lifeExp:
gapminder |> select(-lifeExp)
## # A tibble: 1,704 × 5
## country continent year pop gdpPercap
## <fct> <fct> <int> <int> <dbl>
## 1 Afghanistan Asia 1952 8425333 779.
## 2 Afghanistan Asia 1957 9240934 821.
## 3 Afghanistan Asia 1962 10267083 853.
## 4 Afghanistan Asia 1967 11537966 836.
## 5 Afghanistan Asia 1972 13079460 740.
## 6 Afghanistan Asia 1977 14880372 786.
## 7 Afghanistan Asia 1982 12881816 978.
## 8 Afghanistan Asia 1987 13867957 852.
## 9 Afghanistan Asia 1992 16317921 649.
## 10 Afghanistan Asia 1997 22227415 635.
## # ℹ 1,694 more rowsYou can also rename columns using select(). Follow this pattern: select(old_name = new_name).
gapminder |> select(year, country, life_expectancy = lifeExp)
## # A tibble: 1,704 × 3
## year country life_expectancy
## <int> <fct> <dbl>
## 1 1952 Afghanistan 28.8
## 2 1957 Afghanistan 30.3
## 3 1962 Afghanistan 32.0
## 4 1967 Afghanistan 34.0
## 5 1972 Afghanistan 36.1
## 6 1977 Afghanistan 38.4
## 7 1982 Afghanistan 39.9
## 8 1987 Afghanistan 40.8
## 9 1992 Afghanistan 41.7
## 10 1997 Afghanistan 41.8
## # ℹ 1,694 more rowsAlternatively, there’s a special rename() verb that will, um, rename, while keeping all the other columns:
gapminder |> rename(life_expectancy = lifeExp)
## # A tibble: 1,704 × 6
## country continent year life_expectancy pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779.
## 2 Afghanistan Asia 1957 30.3 9240934 821.
## 3 Afghanistan Asia 1962 32.0 10267083 853.
## 4 Afghanistan Asia 1967 34.0 11537966 836.
## 5 Afghanistan Asia 1972 36.1 13079460 740.
## 6 Afghanistan Asia 1977 38.4 14880372 786.
## 7 Afghanistan Asia 1982 39.9 12881816 978.
## 8 Afghanistan Asia 1987 40.8 13867957 852.
## 9 Afghanistan Asia 1992 41.7 16317921 649.
## 10 Afghanistan Asia 1997 41.8 22227415 635.
## # ℹ 1,694 more rowsArranging data with arrange()
The arrange() verb sorts data. By default it sorts ascendingly, putting the lowest values first:
gapminder |> arrange(lifeExp)
## # A tibble: 1,704 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Rwanda Africa 1992 23.6 7290203 737.
## 2 Afghanistan Asia 1952 28.8 8425333 779.
## 3 Gambia Africa 1952 30 284320 485.
## 4 Angola Africa 1952 30.0 4232095 3521.
## 5 Sierra Leone Africa 1952 30.3 2143249 880.
## 6 Afghanistan Asia 1957 30.3 9240934 821.
## 7 Cambodia Asia 1977 31.2 6978607 525.
## 8 Mozambique Africa 1952 31.3 6446316 469.
## 9 Sierra Leone Africa 1957 31.6 2295678 1004.
## 10 Burkina Faso Africa 1952 32.0 4469979 543.
## # ℹ 1,694 more rowsYou can reverse that by wrapping the column name with desc():
gapminder |> arrange(desc(lifeExp))
## # A tibble: 1,704 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Japan Asia 2007 82.6 127467972 31656.
## 2 Hong Kong, China Asia 2007 82.2 6980412 39725.
## 3 Japan Asia 2002 82 127065841 28605.
## 4 Iceland Europe 2007 81.8 301931 36181.
## 5 Switzerland Europe 2007 81.7 7554661 37506.
## 6 Hong Kong, China Asia 2002 81.5 6762476 30209.
## 7 Australia Oceania 2007 81.2 20434176 34435.
## 8 Spain Europe 2007 80.9 40448191 28821.
## 9 Sweden Europe 2007 80.9 9031088 33860.
## 10 Israel Asia 2007 80.7 6426679 25523.
## # ℹ 1,694 more rowsYou can sort by multiple columns by specifying them in a comma separated list. For example, we can sort by continent and then sort by life expectancy within the continents:
gapminder |>
arrange(continent, desc(lifeExp))
## # A tibble: 1,704 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Reunion Africa 2007 76.4 798094 7670.
## 2 Reunion Africa 2002 75.7 743981 6316.
## 3 Reunion Africa 1997 74.8 684810 6072.
## 4 Libya Africa 2007 74.0 6036914 12057.
## 5 Tunisia Africa 2007 73.9 10276158 7093.
## 6 Reunion Africa 1992 73.6 622191 6101.
## 7 Tunisia Africa 2002 73.0 9770575 5723.
## 8 Mauritius Africa 2007 72.8 1250882 10957.
## 9 Libya Africa 2002 72.7 5368585 9535.
## 10 Algeria Africa 2007 72.3 33333216 6223.
## # ℹ 1,694 more rowsThat’s it!
Those are the main verbs you’ll deal with in this class. There are dozens of other really useful ones—check out the {dplyr} and {tidyr} cheat sheets.
Changing colors, shapes, and sizes, with scale_*()
Recall from session 3 that the grammar of graphics uses a set of layers to define elements of plots:

In tomorrow’s session, you’ll learn all about the Theme layer. Here we’ll briefly cover the Scales layer, which we use for changing aspects of the different aesthetics, like using logged axes or changing colors or shapes.
All the functions that deal with scales conveniently follow the same naming pattern:
scale_AESTHETIC_DETAILS()Here are some common scale functions:
scale_x_continuous()
scale_y_reverse()
scale_color_viridis_c()
scale_shape_manual(values = c(19, 13, 15))
scale_fill_manual(values = c("red", "orange", "blue"))You can see a list of all of the possible scale functions here, and you should reference that documentation (and the excellent examples) often when working with these functions.
As long as you have mapped a variable to an aesthetic with aes(), you can use the scale_*() functions to deal with it. For instance, in this ggplot, we have mapped variables to x, y, and fill, which means we can use those corresponding scale functions to manipulate how those aesthetics are shown. Here we reverse the y-axis (ew, don’t really do this), and we use a discrete viridis color palette:
continent_counts <- gapminder |>
group_by(continent) |>
summarize(countries = n_distinct(country))
ggplot(continent_counts, aes(x = continent, y = countries, fill = continent)) +
geom_col() +
scale_y_reverse() + # lol this is bad; don't do it in real life
scale_fill_viridis_d()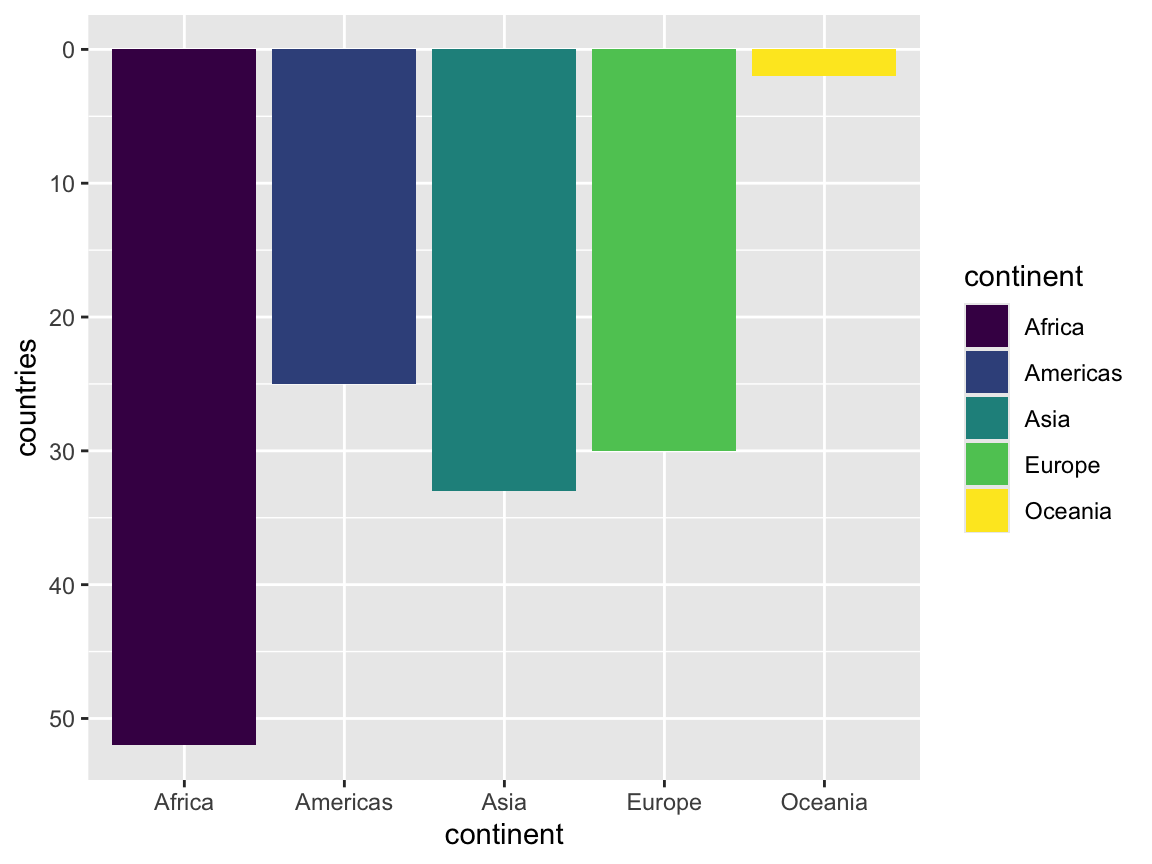
You can also use different arguments in the scale functions—again, check the documentation for examples. For instance, if we want to use the plasma palette from the viridis package, we can set that as an option:
ggplot(continent_counts, aes(x = continent, y = countries, fill = continent)) +
geom_col() +
scale_fill_viridis_d(option = "plasma")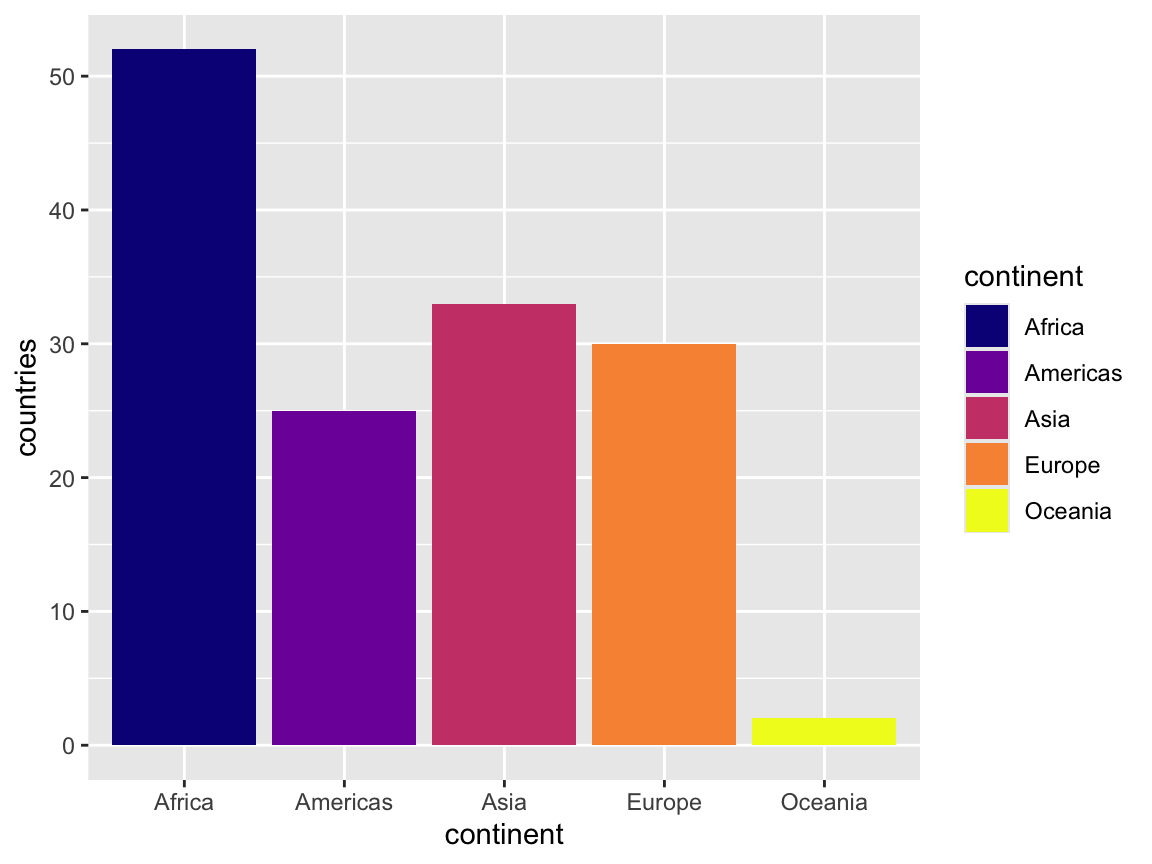
That yellow might be too bright and hard to see, so we can tell ggplot to not use the full range of the palette, ending at 90% of the range instead:
ggplot(continent_counts, aes(x = continent, y = countries, fill = continent)) +
geom_col() +
scale_fill_viridis_d(option = "plasma", end = 0.9)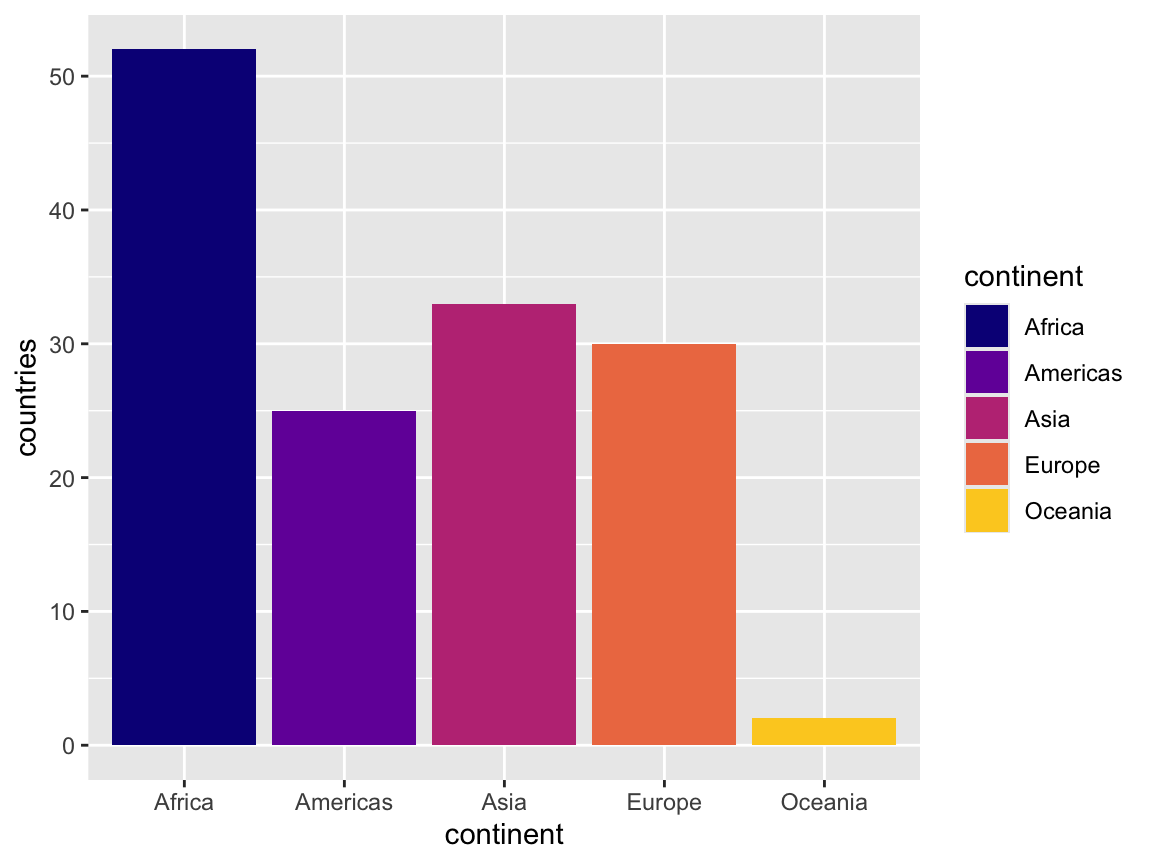
Instead of letting R calculate the colors from a general palette, you can also specify your own colors with scale_fill_manual() and feeding it a list of values—generally as hex codes or a name from a list of built-in R colors:
ggplot(continent_counts, aes(x = continent, y = countries, fill = continent)) +
geom_col() +
scale_fill_manual(values = c("chartreuse4", "cornsilk4", "black", "#fc03b6", "#5c47d6"))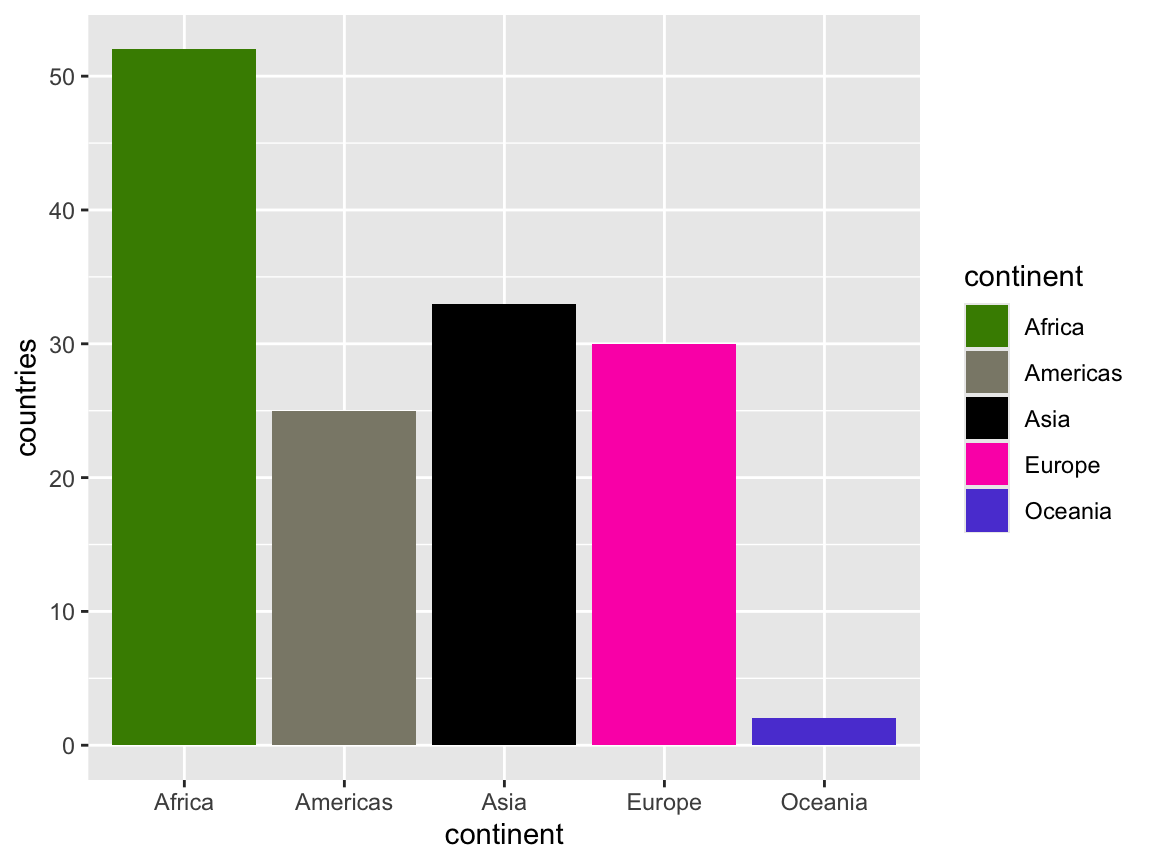
Scale functions also work for other aesthetics like shape or color or size. For instance, consider this plot, which has all three:
gapminder_2007 <- gapminder |>
filter(year == 2007)
ggplot(gapminder_2007,
aes(x = gdpPercap, y = lifeExp,
color = continent, shape = continent, size = pop)) +
geom_point() +
scale_x_log10()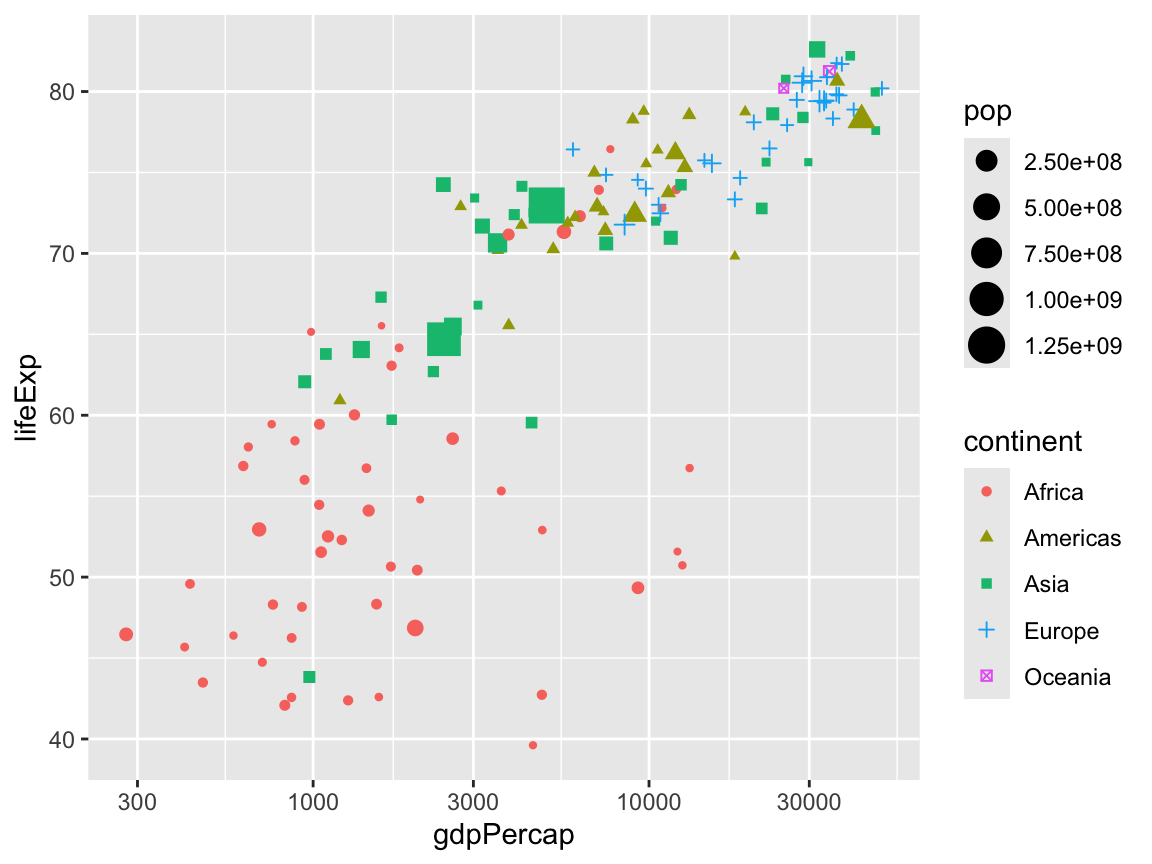
We can change the colors of the points with scale_color_*():
ggplot(gapminder_2007,
aes(x = gdpPercap, y = lifeExp,
color = continent, shape = continent, size = pop)) +
geom_point() +
scale_x_log10() +
scale_color_manual(values = c("chartreuse4", "cornsilk4", "black", "#fc03b6", "#5c47d6"))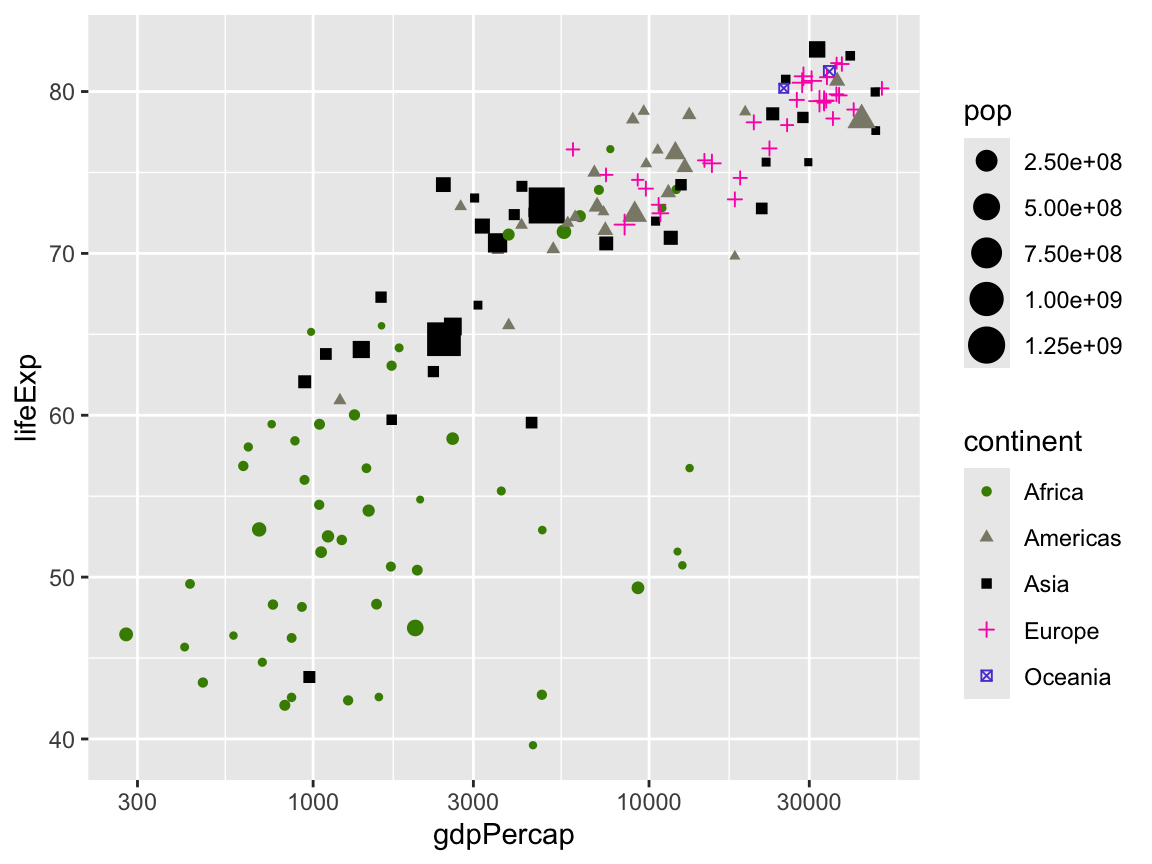
We can change the shapes with scale_shape_*(). If you run ?pch in your console or search for pch in the help, you can see all the possible shapes.
ggplot(gapminder_2007,
aes(x = gdpPercap, y = lifeExp,
color = continent, shape = continent, size = pop)) +
geom_point() +
scale_x_log10() +
scale_shape_manual(values = c(12, 9, 17, 19, 15))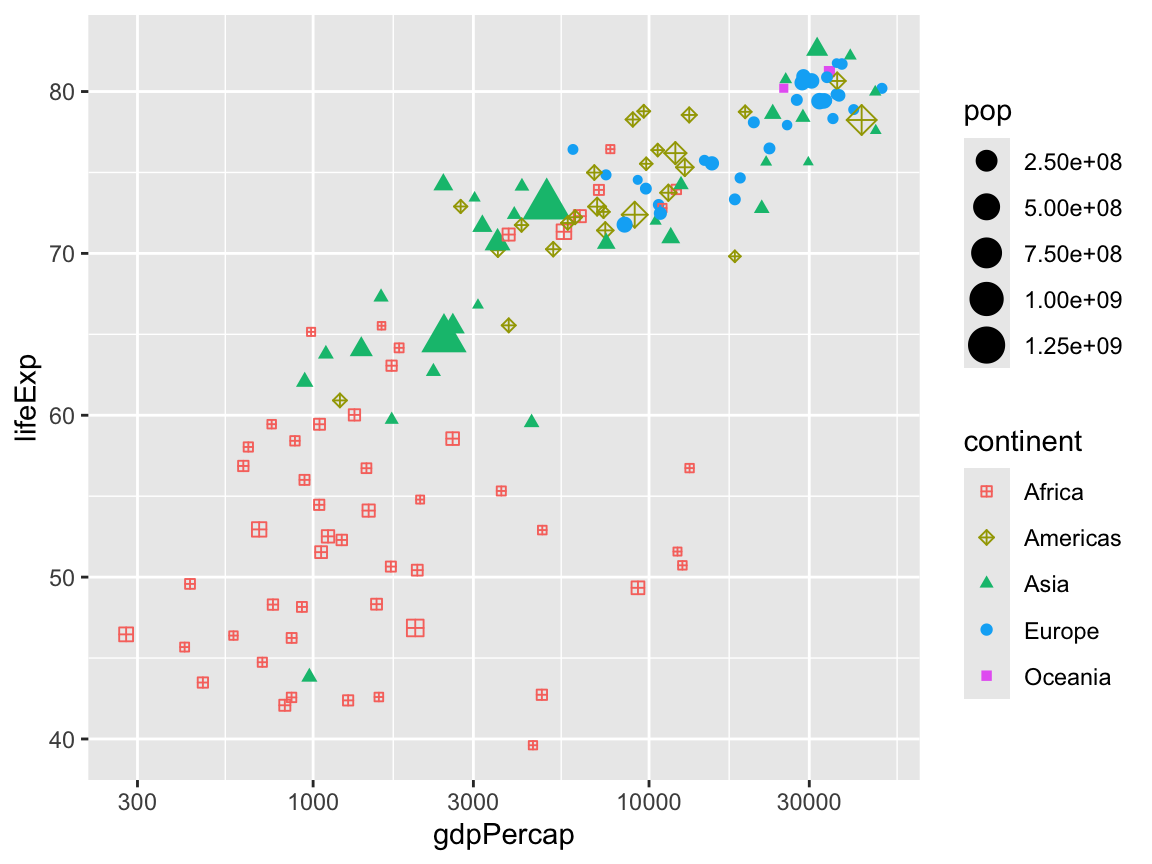
You can change the size with scale_size_*(). Here we make it so the smallest possible size is 1 and the largest is 15:
ggplot(gapminder_2007,
aes(x = gdpPercap, y = lifeExp,
color = continent, shape = continent, size = pop)) +
geom_point() +
scale_x_log10() +
scale_size_continuous(range = c(1, 15))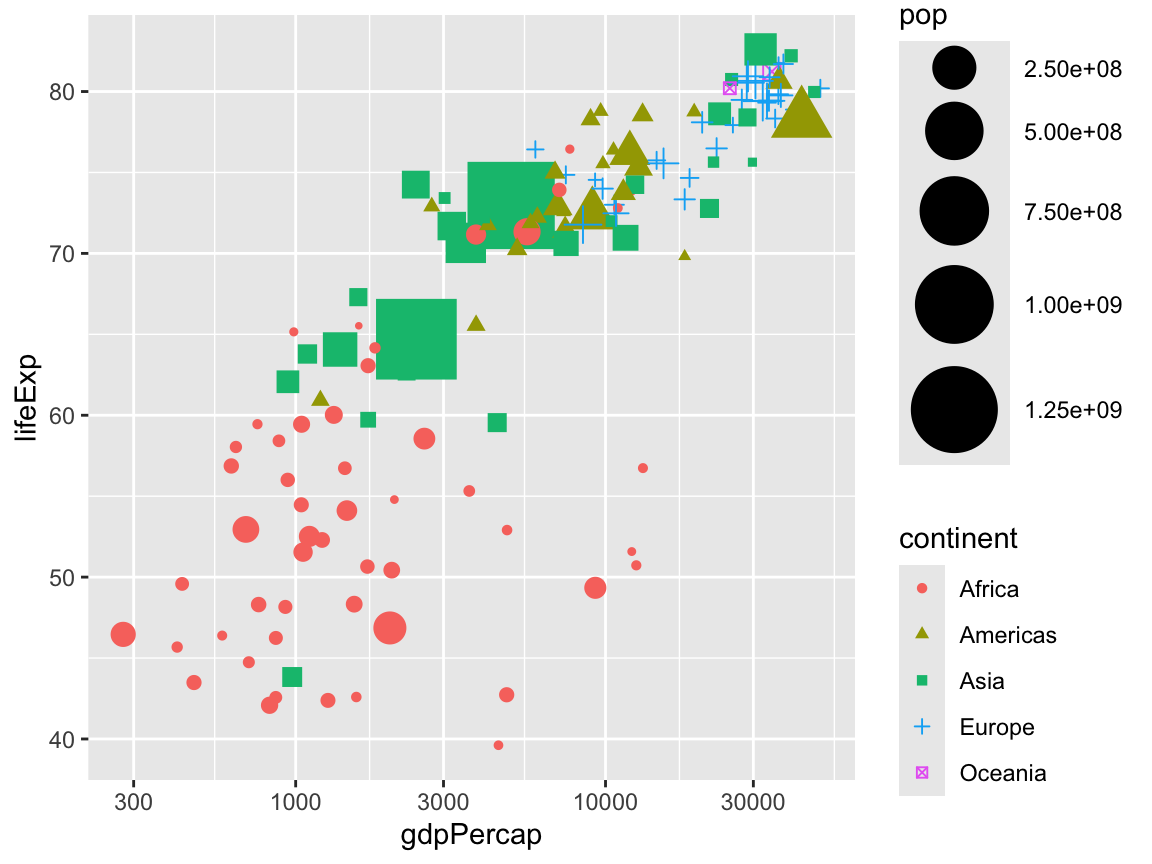
We can even do all three at once:
ggplot(gapminder_2007,
aes(x = gdpPercap, y = lifeExp,
color = continent, shape = continent, size = pop)) +
geom_point() +
scale_x_log10() +
scale_color_manual(values = c("chartreuse4", "cornsilk4", "black", "#fc03b6", "#5c47d6")) +
scale_shape_manual(values = c(12, 9, 17, 19, 15)) +
scale_size_continuous(range = c(1, 15))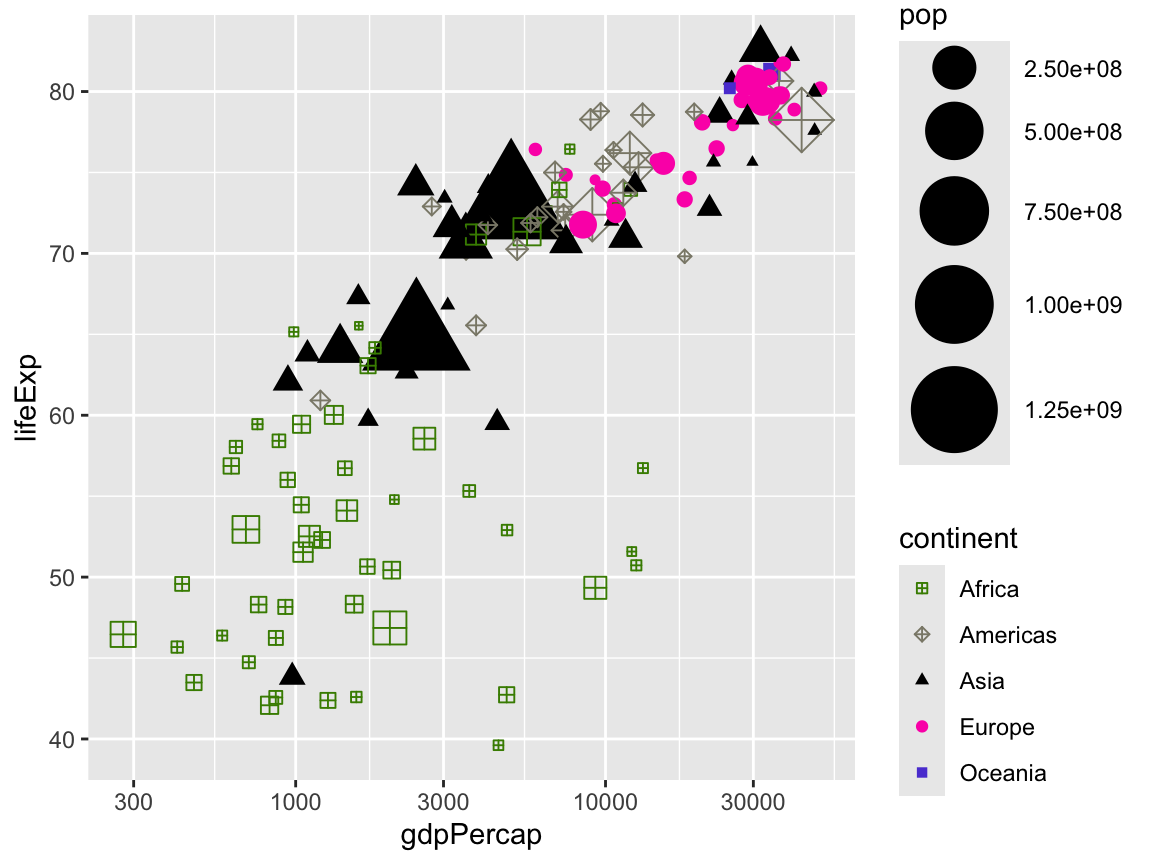
Phew. That’s ugly.
One last thing we can do with scales is format how they show up on the plot. Notice how the population legend uses scientific notation like 2.50e+08. This means you need to move the decimal point 8 places to the right, making it 250000000. Leaving it in scientific notation isn’t great because it makes it really hard to read and interpret.
If you load the {scales} library (which is installed as part of {tidyverse} but isn’t automatically loaded), you can use some neat helper functions to reformat the text that shows up in plots. For instance, we can make it so population is formatted as a number with commas every 3 numbers, and the x-axis is formatted as dollars:
library(scales)
ggplot(gapminder_2007,
aes(x = gdpPercap, y = lifeExp,
color = continent, shape = continent, size = pop)) +
geom_point() +
scale_x_log10(labels = label_dollar()) +
scale_size_continuous(labels = label_comma())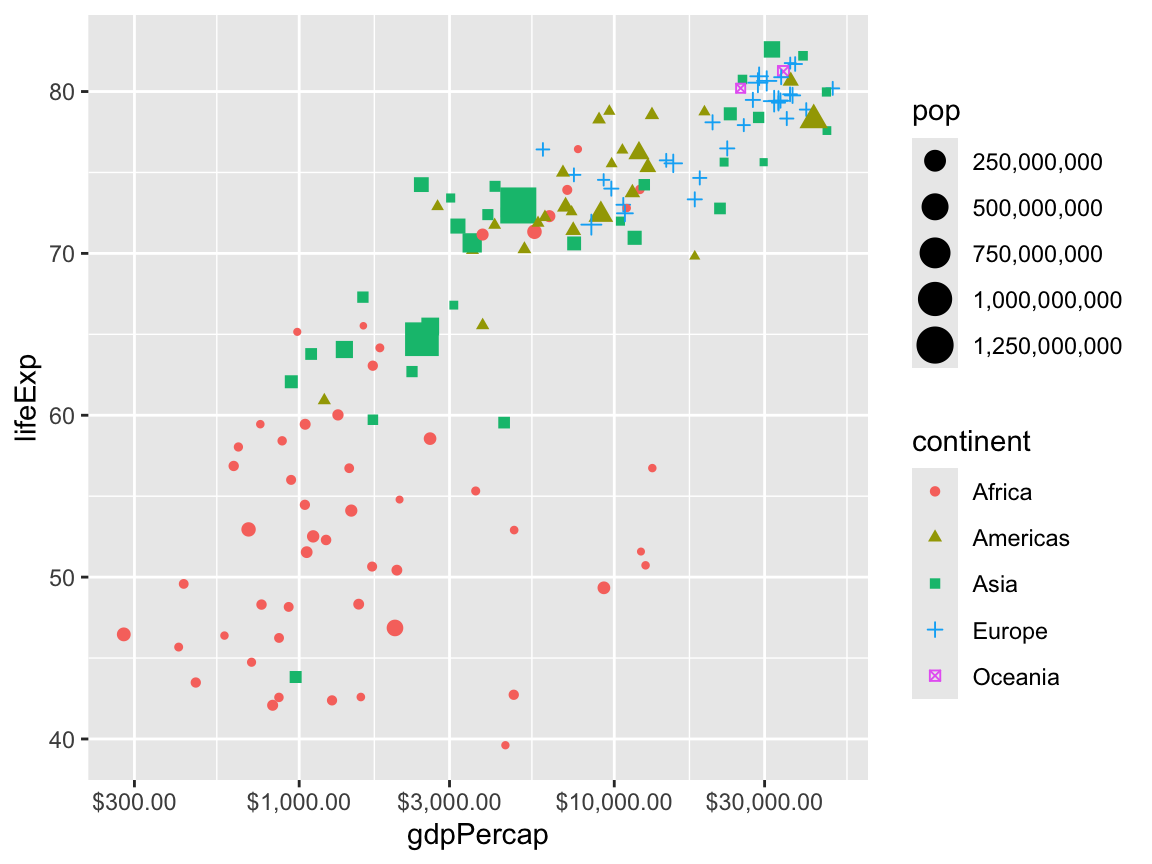
Check the documentation for {scales} for details about all the labeling functions it has, including dates, percentages, p-values, LaTeX math, etc.